Biomed Res Bull. 2(2):70-74.
doi: 10.34172/biomedrb.2024.11
Original Article
Investigating the Expression of Long Non-coding RNA Gene BANCR in Tumor Tissues and Adjacent Non-tumor Tissues of Patients with Breast Cancer
Raha Nikanfar 1  , Alireza Nikanfar 1, Mahsa Nikanfar 1, *
, Alireza Nikanfar 1, Mahsa Nikanfar 1, * 
Author information:
1Hematology and Oncology Research Center, Tabriz, University of Medical Sciences, Tabriz, Iran
Abstract
Background:
Breast cancer is one of the most common types of cancer worldwide, and long non-coding RNAs (lncRNAs) have been implicated in its development and progression.
Methods:
In this study, we investigated the expression of BRAF-activated non-coding RNA (BANCR) in tumor tissues and adjacent non-tumor tissues of 123 breast cancer patients.
Results:
Our results showed that the expression of BANCR was significantly higher in tumor tissues compared to adjacent non-tumor tissues. Based on the results, the mean fold change was 7.894 (P<0.001). Moreover, the result revealed that the expression of BANCR was increased in patients with lymph node invasion in comparison to patients without lymph node invasion (LogFC=1.358, P<0.001).
Conclusion:
The study findings suggest that BANCR may be a potential biomarker for breast cancer; however, further research is needed to confirm these results.
Keywords: Breast cancer, LncRNA, BANCR, Lymph node invasion
Copyright and License Information
© 2024 The Author(s).
This is an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This study was self-funded by the authors and received no external financial support from any funding organization.
Introduction
Breast cancer is the most commonly diagnosed cancer accounting for approximately 30% of all cancer cases in women worldwide and the leading cause of cancer-related deaths in women worldwide.1 Despite advancements in early detection and treatment, the molecular mechanisms underlying breast cancer pathogenesis and progression remain elusive.2 Despite significant advances in diagnosis and treatment, breast cancer remains a major health concern, with high morbidity and mortality rates.3 Therefore, it is important to investigate the underlying molecular mechanisms that contribute to the development and progression of breast cancer.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules that are longer than 200 nucleotides and do not code for proteins.4 LncRNAs have been shown to play critical roles in various biological processes, including gene regulation, chromatin modification, and epigenetic regulation.5 Emerging evidence suggests that lncRNAs are involved in the pathogenesis of breast cancer, and dysregulation of lncRNA expression has been associated with tumor initiation, progression, and metastasis.6
One lncRNA that has gained attention in breast cancer research is BRAF-activated non-coding RNA (BANCR). BANCR was first identified in melanoma and has been shown to be dysregulated in various cancers, including breast cancer.7 BANCR has been implicated in cancer cell proliferation, invasion, and metastasis, suggesting that it may play a critical role in breast cancer progression.8
In this study, we aimed to investigate the expression of BANCR in tumor tissues and adjacent non-tumor tissues of patients with breast cancer. We hypothesized that BANCR expression would be upregulated in breast cancer tissues compared to adjacent non-tumor tissues and that its expression would correlate with the clinicopathological features such as lymph node invasion. To test our hypothesis, we collected breast cancer tissues and adjacent non-tumor tissues from patients who underwent surgery for breast cancer. We analyzed BANCR expression levels using quantitative real-time PCR and correlated the results with clinicopathological features of the patients. The findings of this study have the potential to provide insights into the role of BANCR in breast cancer and may contribute to the development of new diagnostic and therapeutic strategies for this disease.
Materials and Methods
Collection of Patient Samples
The study enrolled 123 female patients who were diagnosed with breast cancer and underwent surgical resection at Nournejat hospital. Patients with a history of chemotherapy, radiation therapy, or hormone therapy were excluded from the study. Tumor tissues and adjacent non-tumor tissues were collected immediately after surgery and snap-frozen in liquid nitrogen. Then, the samples were transferred to a -80 °C freezer until RNA extraction. The demographic data of these 123 patients are summarized in Table 1.
Table 1.
Demographic Data of Patients
|
|
Parameters
|
Patients (%)
|
| Age |
Under 50 |
68 (55%) |
| Above 50 |
55 (45%) |
| Pathology |
Invasive ductal carcinoma |
83 (67%) |
| Invasive lobular carcinoma |
40 (33%) |
| Stage |
I and II |
39 (31%) |
| III and IV |
84 (69%) |
| Lymph node invasion |
Yes |
58 (47%) |
| No |
65 (53%) |
RNA Extraction
Total RNA was extracted from the frozen tissue samples using the TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Briefly, 1 mL of TRIzol reagent was added per 100 mg of tissue, followed by homogenization using a tissue homogenizer. After phase separation with chloroform, RNA was precipitated using isopropanol, washed with 75% ethanol, and dissolved in RNase-free water. The quantity and purity of the extracted RNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
RNA Extraction Quality Control
The quality of the extracted RNA was evaluated using the NanoDrop spectrophotometer, which measures the absorbance of RNA at wavelengths of 260, 280, and 230 nm. The A260/A280 ratio indicates the purity of RNA, and a value between 1.8 and 2.0 indicates pure RNA without contamination from proteins or other contaminants. The A260/A230 ratio reflects the presence of contaminants such as organic compounds or salt, and a value above 2.0 indicates that the RNA sample is free from contaminants.
cDNA Synthesis
Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol. Briefly, 1 μg of RNA was mixed with 2 μL of 10x RT buffer, 0.8 μL of 25x dNTP mix, 2 μL of 10x RT random primers, 1 μL of MultiScribeTM Reverse Transcriptase, and 4.2 μL of nuclease-free water. The reaction mixture was incubated at 25°C for 10 minutes, followed by 37 °C for 120 minutes and 85 °C for 5 minutes.
Primer Design
Primers for BANCR and the reference gene (B actin) were designed using Primer3 software version 4.1.0. The primer sequences were as follows: BANCR forward primer, 5’- ACAGGACTCCATGGCAAACG -3’; BANCR reverse primer, 5’- ATGAAGAAAGCCTGGTGCAGT -3’; B actin forward primer, 5’- AGAGCTACGAGCTGCCTGAC -3’; B actin reverse primer, 5’- AGCACTGTGTTGGCGTACAG -3’.
Real-Time Polymerase Chain Reaction
Real-time polymerase chain reaction (PCR) was performed using the PowerUp SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) on the QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The reaction mixture consisted of 10 μL of PowerUp SYBR Green Master Mix, 0.5 μL of each primer (10 μM), 2 μL of cDNA template, and 7 μL of nuclease-free water. The thermal cycling conditions were as follows: initial denaturation at 95 °C for 10 minutes, followed by 40 cycles of denaturation at 95 °C for 15 seconds, and annealing and extension at 60 °C for 60 seconds. Melting curve analysis was performed to verify the specificity of the PCR products.
Statistical Analysis
Statistical analysis was performed using SPSS version 23.0. The expression levels of BANCR were calculated using the 2-ΔΔCt method and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. The data were presented as mean ± standard deviation (SD). Differences in BANCR expression levels between tumor tissues and adjacent non-tumor tissues were analyzed using paired t test. Correlations between BANCR expression and clinicopathological parameters were assessed using Pearson correlation analysis. P values < 0.05 were considered statistically significant.
Results
Expression of BANCR in Tumor Tissues and Adjacent Non-tumor Tissues
The expression of BANCR was evaluated in tumor tissues and adjacent non-tumor tissues of breast cancer patients using real-time quantitative PCR. The expression level of BANCR was normalized to that of GAPDH, and the relative expression levels were calculated using the 2-ΔΔCt method.
The expression level of BANCR was significantly higher in tumor tissues than in adjacent non-tumor tissues (mean fold change, 7.894, P < 0.001) (Figure 1).
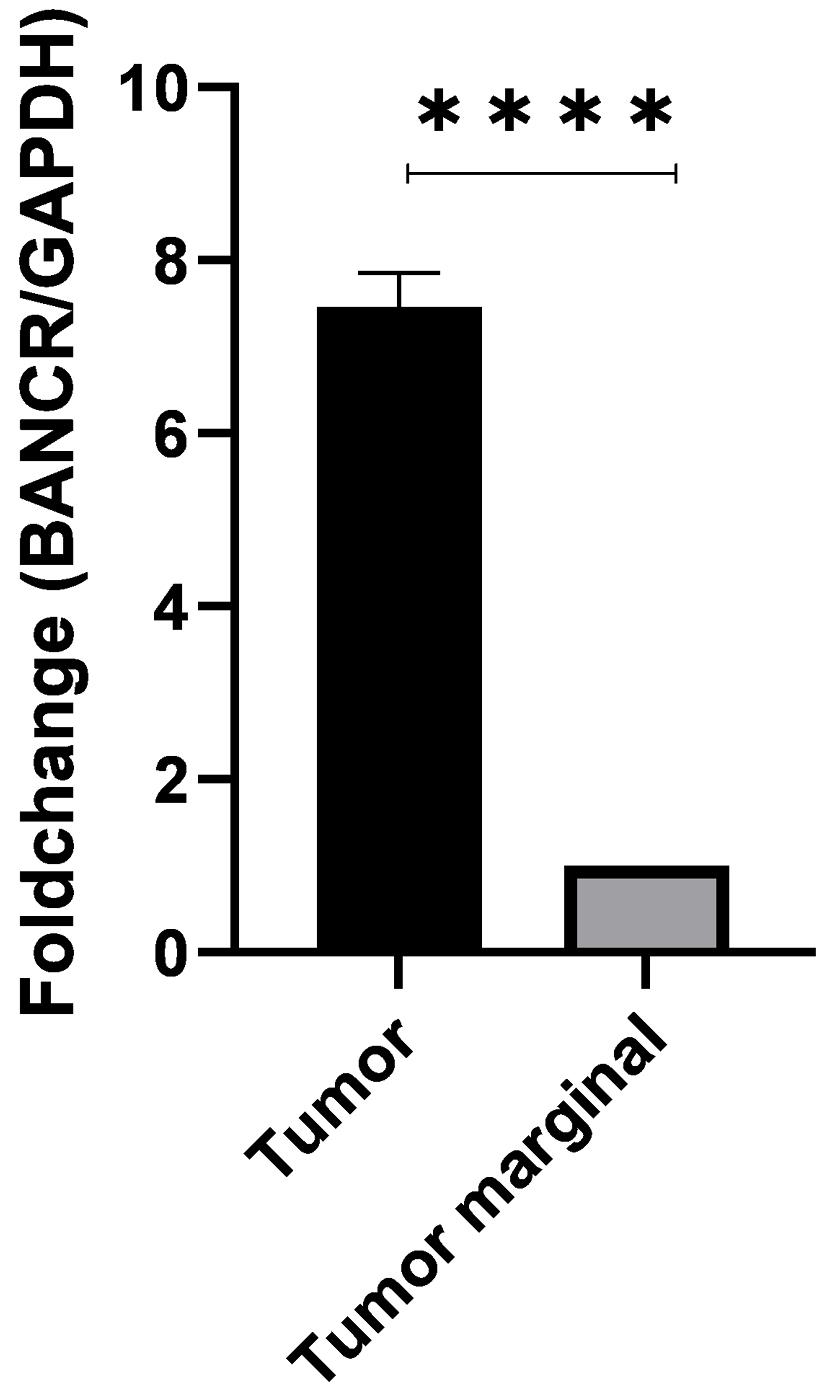
Figure 1.
Expression of BANCR in Tumor Tissues and Adjacent Non-tumor Tissues
.
Expression of BANCR in Tumor Tissues and Adjacent Non-tumor Tissues
Expression of BANCR in Patients With and without Lymph Node Invasion
To investigate the potential association of BANCR expression with lymph node invasion, we compared the expression levels of BANCR in tumor tissue samples obtained from breast cancer patients with and without lymph node invasion. Using quantitative real-time PCR, we measured the expression levels of BANCR and calculated the log fold change (logFC) between the two groups.
Our results show that BANCR expression was significantly upregulated in breast cancer patients with lymph node invasion compared to patients without lymph node invasion. The logFC expression of BANCR in these patients was 1.358 times higher than in patients without lymph node invasion (P < 0.001). These findings suggest that BANCR may play a role in promoting lymph node invasion in patients with breast cancer (Figure 2).
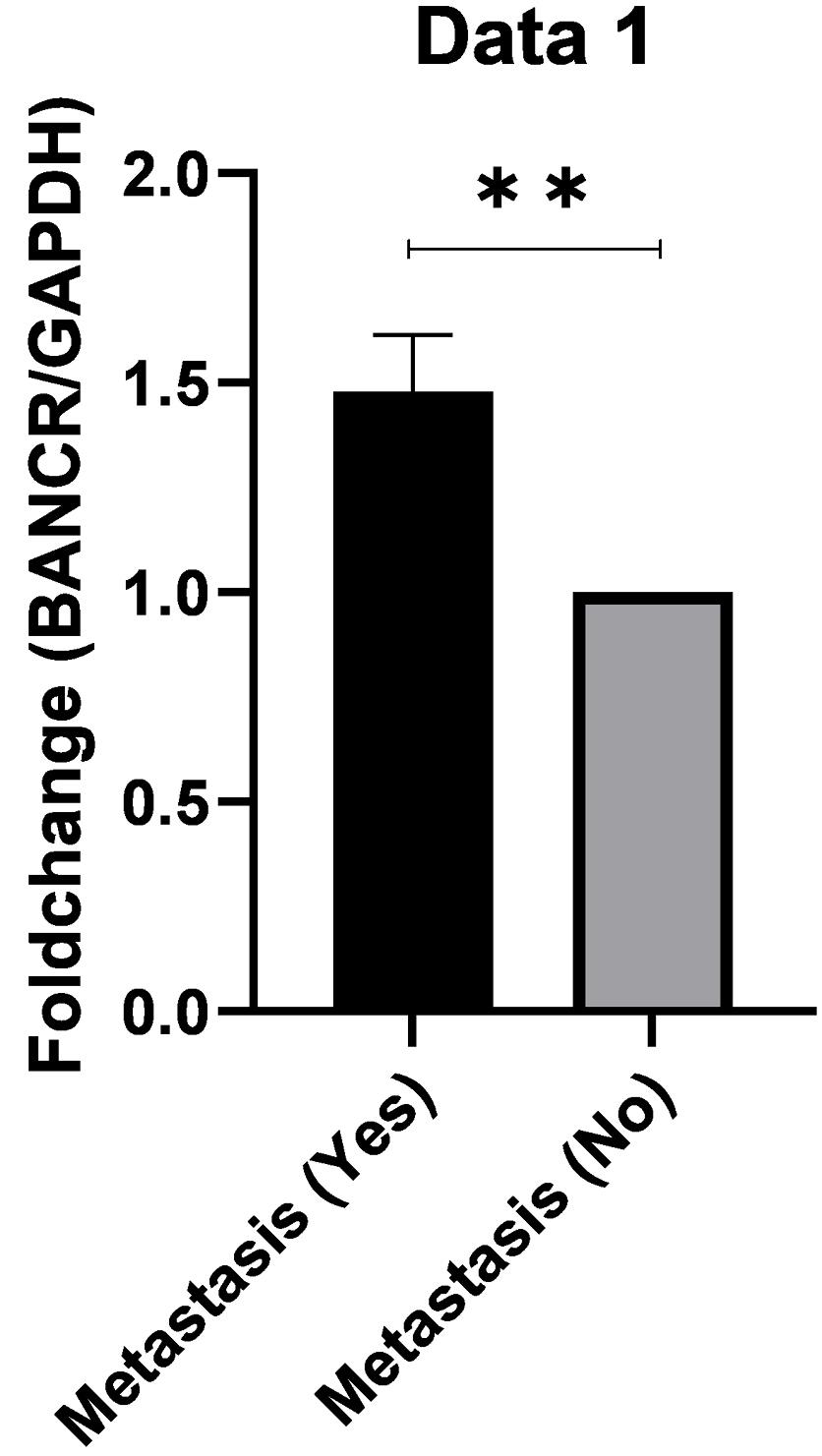
Figure 2.
Expression of BANCR in Patients with and without Lymph Node Invasion
.
Expression of BANCR in Patients with and without Lymph Node Invasion
Discussion
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer-related deaths among women worldwide.9 Despite significant advances in early detection and treatment, breast cancer remains a major public health concern, highlighting the need for continued research efforts to identify novel biomarkers and therapeutic targets for this disease.10
LncRNAs have emerged as a new class of functional RNA molecules that play critical roles in various biological processes, including gene regulation, chromatin modification, and epigenetic regulation.11 Dysregulation of lncRNAs has been implicated in various diseases, including cancer.12 BANCR is a recently identified lncRNA that has been shown to be upregulated in various types of cancer, including melanoma, colorectal cancer, and hepatocellular carcinoma.13,14 However, the role of BANCR in breast cancer remains unclear.
In this study, we investigated the expression of BANCR in tumor tissues and adjacent non-tumor tissues of breast cancer patients and found that BANCR was significantly upregulated in tumor tissues compared with adjacent non-tumor tissues. This result is consistent with previous studies that have shown the upregulation of BANCR in various types of cancer. Furthermore, we found that the expression of BANCR was higher in patients with lymph node invasion compared with patients without lymph node invasion, suggesting a potential role of BANCR in the development of breast cancer and lymph node invasion.
The precise mechanisms underlying the role of BANCR in breast cancer are not well understood. However, several studies have provided insights into the potential molecular mechanisms by which BANCR may contribute to the development and progression of breast cancer. For example, BANCR has been shown to promote cell proliferation, invasion, and migration in various cancer cell lines. In papillary thyroid carcinoma, BANCR has been reported to regulate the expression of several genes involved in cancer cell proliferation and migration, such as E-cadherin, N-cadherin, and vimentin. In addition, BANCR has been shown to interact with several proteins, including EZH2 and SUZ12, which are involved in epigenetic regulation.15 These findings suggest that BANCR may contribute to the development and progression of breast cancer by regulating gene expression and epigenetic modifications.
Furthermore, our results showed that the expression of BANCR was higher in patients with lymph node invasion compared with patients without lymph node invasion, indicating that BANCR may be involved in the aggressiveness and invasiveness of breast cancer. Previous studies have also shown a correlation between BANCR expression and poor prognosis in various types of cancer. For example, Jiang et al conducted a study to investigate the expression level and functional role of the lncRNA BANCR in breast cancer. Their findings revealed that BANCR was overexpressed in breast cancer cell lines and tissues. The elevated expression of BANCR was associated with clinical indicators of aggressive disease, including larger tumor size, lymph node metastasis, and advanced tumor-node-metastasis stage. Moreover, high expression level of BANCR correlated with poor overall survival and early recurrence in breast cancer patients. Functional experiments using BANCR knockdown and overexpression demonstrated its involvement in promoting breast cancer cell proliferation, migration, and invasion.16 In melanoma, BANCR has been shown to promote metastasis by regulating the expression of genes involved in cell migration and invasion.17 These findings suggest that BANCR may be a potential prognostic marker and therapeutic target for breast cancer.
In conclusion, our study provides evidence that BANCR is upregulated in tumor tissues of breast cancer patients and is associated with clinicopathological characteristics of the patients such as metastasis. Although our study has several limitations, including a relatively small sample size and lack of functional studies to elucidate the precise mechanisms underlying the role of BANCR in breast cancer, our findings suggest that BANCR may be a potential biomarker and therapeutic target for breast cancer. Further studies are warranted to validate our findings and to investigate the potential clinical implications of BANCR in breast cancer. Moreover, future studies may explore the potential use of BANCR as a diagnostic or prognostic tool, as well as a therapeutic target for breast cancer.
One potential avenue for future research is to investigate the role of BANCR in breast cancer cell lines and animal models. Functional studies can provide insights into the molecular mechanisms underlying the role of BANCR in breast cancer and may identify potential therapeutic targets for this disease. For example, targeting the expression of BANCR or its downstream effectors may represent a novel approach for the treatment of breast cancer.
Another important area for future research is to investigate the potential use of BANCR as a diagnostic or prognostic biomarker for breast cancer. Previous studies have shown that BANCR expression is associated with clinicopathological characteristics and prognosis in various types of cancer. In breast cancer, BANCR may serve as a potential biomarker for early detection or prediction of disease progression. Moreover, the development of non-invasive methods for detecting BANCR expression, such as circulating tumor cells or exosomes, may provide a promising avenue for clinical applications.
Finally, future studies may investigate the potential use of BANCR as a therapeutic target for breast cancer. Several strategies can be employed to target the expression of BANCR or its downstream effectors, such as small molecule inhibitors or RNA interference. For example, previous studies have shown that inhibition of BANCR expression can reduce cell proliferation and migration in various cancer cell lines.18 Furthermore, targeting BANCR-regulated genes or downstream effectors, such as EZH2 or SUZ12, may represent a potential therapeutic strategy for breast cancer.
Conclusion
In summary, our study provides evidence that BANCR is upregulated in tumor tissues of breast cancer patients and is associated with clinicopathological characteristics of the patients including metastasis. Although our findings warrant further validation and functional studies, they suggest that BANCR may be a potential biomarker and therapeutic target for breast cancer. Further research efforts are needed to elucidate the precise mechanisms underlying the role of BANCR in breast cancer and to explore its potential clinical applications. The identification of novel biomarkers and therapeutic targets is essential for improving the diagnosis, prognosis, and treatment of breast cancer, and BANCR represents a promising avenue for future research in this area.
Authors’ Contribution
Conceptualization: Mahsa Nikanfar.
Data curation: Alireza Nikanfar.
Formal analysis: Raha Nikanfar.
Funding acquisition: Mahsa Nikanfar.
Investigation: Raha Nikanfar.
Methodology: Alireza Nikanfar,Raha Nikanfar.
Project administration: Mahsa Nikanfar.
Resources: Alireza Nikanfar.
Software: Raha Nikanfar.
Supervision: Mahsa Nikanfar.
Validation: Alireza Nikanfar.
Visualization: Raha Nikanfar.
Writing–original draft: Raha Nikanfar.
Writing–review & editing: Mahsa Nikanfar.
Competing Interests
None.
Ethical Approval
This study was approved by Shahid Beheshti University of Medical Sciences (Ethical code: IR.SBMU.MSP.REC.1399.474).
References
- Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021; 134(7):783-91. doi: 10.1097/cm9.0000000000001474 [Crossref] [ Google Scholar]
- Suva LJ, Griffin RJ, Makhoul I. Mechanisms of bone metastases of breast cancer. Endocr Relat Cancer 2009; 16(3):703-13. doi: 10.1677/erc-09-0012 [Crossref] [ Google Scholar]
- Anderson BO, Ilbawi AM, El Saghir NS. Breast cancer in low- and middle-income countries (LMICs): a shifting tide in global health. Breast J 2015; 21(1):111-8. doi: 10.1111/tbj.12357 [Crossref] [ Google Scholar]
- Hobuß L, Bär C, Thum T. Long non-coding RNAs: at the heart of cardiac dysfunction?. Front Physiol 2019; 10:30. doi: 10.3389/fphys.2019.00030 [Crossref] [ Google Scholar]
- Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J. LncRNA structural characteristics in epigenetic regulation. Int J Mol Sci 2017; 18(12):2659. doi: 10.3390/ijms18122659 [Crossref] [ Google Scholar]
- Amelio I, Bernassola F, Candi E. Emerging roles of long non-coding RNAs in breast cancer biology and management. Semin Cancer Biol 2021; 72:36-45. doi: 10.1016/j.semcancer.2020.06.019 [Crossref] [ Google Scholar]
- Choudhari R, Sedano MJ, Harrison AL, Subramani R, Lin KY, Ramos EI. Long noncoding RNAs in cancer: from discovery to therapeutic targets. Adv Clin Chem 2020; 95:105-47. doi: 10.1016/bs.acc.2019.08.003 [Crossref] [ Google Scholar]
- Zhou S, He Y, Yang S, Hu J, Zhang Q, Chen W. The regulatory roles of lncRNAs in the process of breast cancer invasion and metastasis. Biosci Rep 2018; 38(5):BSR20180772. doi: 10.1042/bsr20180772 [Crossref] [ Google Scholar]
- Houghton SC, Hankinson SE. Cancer progress and priorities: breast cancer. Cancer Epidemiol Biomarkers Prev 2021; 30(5):822-44. doi: 10.1158/1055-9965.Epi-20-1193 [Crossref] [ Google Scholar]
- Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res 2013; 15(5):R92. doi: 10.1186/bcr3493 [Crossref] [ Google Scholar]
- Costa FF. Non-coding RNAs: meet thy masters. Bioessays 2010; 32(7):599-608. doi: 10.1002/bies.200900112 [Crossref] [ Google Scholar]
- Wang J, Ye C, Xiong H, Shen Y, Lu Y, Zhou J. Dysregulation of long non-coding RNA in breast cancer: an overview of mechanism and clinical implication. Oncotarget 2017; 8(3):5508-22. doi: 10.18632/oncotarget.12537 [Crossref] [ Google Scholar]
- Zhou T, Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World J Surg Oncol 2016; 14(1):8. doi: 10.1186/s12957-015-0757-5 [Crossref] [ Google Scholar]
- Zou Y, Li J, Chen Y, Xiao H, Zhang F, Yu D. BANCR: a novel oncogenic long non-coding RNA in human cancers. Oncotarget 2017; 8(55):94997-5004. doi: 10.18632/oncotarget.22031 [Crossref] [ Google Scholar]
- Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 2014; 13:68. doi: 10.1186/1476-4598-13-68 [Crossref] [ Google Scholar]
- Jiang J, Shi SH, Li XJ, Sun L, Ge QD, Li C. Long non-coding RNA BRAF-regulated lncRNA 1 promotes lymph node invasion, metastasis and proliferation, and predicts poor prognosis in breast cancer. Oncol Lett 2018; 15(6):9543-52. doi: 10.3892/ol.2018.8513 [Crossref] [ Google Scholar]
- Melixetian M, Pelicci PG, Lanfrancone L. Regulation of LncRNAs in melanoma and their functional roles in the metastatic process. Cells 2022; 11(3):577. doi: 10.3390/cells11030577 [Crossref] [ Google Scholar]
- Jiang W, Zhang D, Xu B, Wu Z, Liu S, Zhang L. Long non-coding RNA BANCR promotes proliferation and migration of lung carcinoma via MAPK pathways. Biomed Pharmacother 2015; 69:90-5. doi: 10.1016/j.biopha.2014.11.027 [Crossref] [ Google Scholar]