Biomed Res Bull. 2(3):124-132.
doi: 10.34172/biomedrb.2024.19
Original Article
Investigating the Effects of Thiosemicarbazone Complexes and Chemotherapy Drugs on the Expression Profiles of ANRIL, CCAT1, NEAT1, and GAS5 lncRNAs in the Jurkat E6.1 Acute Lymphoblastic Leukemia Cell Line
Saedeh Hatampour 1, #  , Mahsa Qaed 1, #
, Mahsa Qaed 1, #  , Neda Zahmatkesh 1, Sina Mirzaahmadi 1, Golnaz Asaadi Tehrani 1, 2, *
, Neda Zahmatkesh 1, Sina Mirzaahmadi 1, Golnaz Asaadi Tehrani 1, 2, * 
Author information:
1Molecular Genetic Department of Genetics, Zanjan Branch, Islamic Azad University, Zanjan, Iran
2Notre Dame University, IN, USA
#Equally contributed.
Abstract
Background:
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease and one of the most common malignancies in children, treated with chemotherapy drugs and thiosemicarbazone complexes. This study investigated the relationship between changes in the expression of nuclear enriched abundant transcript 1 (NEAT1), growth arrest-specific 5 (GAS5), antisense non-coding RNA in the INK4 locus (ANRIL), and colon cancer-associated transcript 1 (CCAT1) long non-coding RNAs (lncRNAs) in the Jurkat E6.1 ALL cell line treated with chemotherapy drugs and thiosemicarbazone complexes.
Methods:
The Jurkat E6.1 ALL cell line was treated with chemotherapy drugs and thiosemicarbazone complexes prepared in different concentrations for different periods of time (24, 48, and 72 hours). RNA extraction and complementary DNA synthesis were performed, and the expression levels of NEAT1, GAS5, ANRIL, and CCAT1 lncRNAs were examined by real-time polymerase chain reaction. The obtained results were statistically analyzed using REST software.
Results:
Changes in GAS5 expression were observed at various concentrations of drugs after different time periods. The highest significant increase in expression was observed in methotrexate at a concentration of 10 μM in 24 hours. Changes in NEAT1 expression were time-dependent, and the most significant decrease was found at 72 hours with Arac at a concentration of 5 μM. Changes in CCAT1 gene expression were noticeable for all drug compounds and concentrations at different time periods. Changes in ANRIL expression were time-dependent, and the most significant decrease was observed at 72 hours with Complex 3.
Conclusion:
The findings revealed that changes in the expression of NEAT1, GAS5, ANRIL, and CCAT1 lncRNAs were affected by the treatment with chemotherapy drugs and thiosemicarbazone complexes in the Jurkat E6.1 ALL cell line. The results also indicated that all drug compounds and concentrations were time-dependent in terms of efficacy. These findings may contribute to the development of new therapeutic strategies for ALL treatment.
Keywords: Acute lymphoblastic leukemia, LncRNA, Chemotherapeutic agents, Thiosemicarbazone complexes
Copyright and License Information
© 2024 The Author(s).
This is an open access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Funding Statement
This study was self-funded by the authors and received no external financial support from any funding organization.
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous malignancy characterized by the uncontrolled proliferation of lymphoblasts, predominantly affecting children.1 It is the most common form of cancer in pediatric patients, accounting for approximately 30% of childhood malignancies.2 Despite significant advancements in the treatment of ALL, including the use of chemotherapy drugs and targeted therapies, the disease still poses significant challenges due to its complex and diverse nature.
Long non-coding RNAs (lncRNAs) have recently emerged as key players in the regulation of gene expression and cellular processes, including cancer development and progression.3,4 These non-protein-coding transcripts have been implicated in diverse biological functions, such as chromatin remodeling, transcriptional regulation, and post-transcriptional processing.5 In the context of ALL, exploring the role of lncRNAs can provide valuable insights into the underlying molecular mechanisms and potentially identify novel therapeutic targets.6,7
Chemotherapy drugs and thiosemicarbazone complexes have been pivotal in the treatment of ALL, aiming to eradicate leukemic cells and achieve disease remission.8 However, the specific impact of these treatment modalities on the expression of lncRNAs in ALL remains poorly understood. Therefore, this study seeks to investigate the relationship between changes in the expression of nuclear enriched abundant transcript 1 (NEAT1), growth arrest-specific 5 (GAS5), antisense non-coding RNA in the INK4 locus (ANRIL), and colon cancer-associated transcript 1 (CCAT1) lncRNAs in the Jurkat E6.1 ALL cell line following exposure to chemotherapy drugs and thiosemicarbazone complexes.
The Jurkat E6.1 cell line, derived from a T-cell ALL patient, serves as a valuable in vitro model for studying ALL pathogenesis and evaluating therapeutic interventions. In this study, the cell line was treated with various concentrations of chemotherapy drugs and thiosemicarbazone complexes and prepared according to established protocols. Different concentrations and treatment durations (24, 48, and 72 hours) have been considered to capture the dynamic changes in lncRNA expression over time and assess the dose-dependent effects of the treatments. Understanding the intricate regulatory networks involving lncRNAs in ALL pathogenesis and treatment response is of utmost importance for the development of novel therapeutic strategies. It is expected that the findings of this study contribute to our knowledge of lncRNA involvement in ALL and provide a basis for further investigation into the functional significance of NEAT1, GAS5, ANRIL, and CCAT1 lncRNAs in ALL. Furthermore, elucidating the mechanistic links between lncRNAs and the efficacy of chemotherapy drugs and thiosemicarbazone complexes may pave the way for personalized treatment approaches and the identification of novel therapeutic targets. A comprehensive understanding of the regulatory roles of these lncRNAs in ALL can provide crucial insights into the underlying molecular mechanisms driving leukemic cell growth and survival.
Materials and Methods
The case-control study was conducted at the Research Science Center of the Islamic Azad University of Zanjan from April to September 2019. Acute Jurkat E6.1 human lymphoblastic leukemia cells with 80% density (2 × 105 cells/cm2) were obtained from the Pasteur Institute of Iran in Passage 1 for the study.
Cell Culture and Preparation of Different Concentrations of Drug
The Jurkat E6.1 ALL cell was first grown in Dulbecco’s modified Eagle’s medium (RPMI 90%) with 10% fetal bovine serum for six days at 37 °C with 5% CO2. The cells were then passaged every two days, with each passage involving the insertion of the cells into a flask containing new growth media. The cells from the fourth passage were chosen for the study, counted, and stained with Trypan blue. The number of cells per cm2 was calculated to be 104 × 3 cells/cm2. Next, the cells were categorized into the control and sample groups.
The cytotoxicity and half-maximal inhibitory concentration (IC50) value of thiosemicarbazone Ni/Cu complexes were determined using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay. The combining CuHL1 after 24, 48, and 72 hours resulted in 20 ± 2.5, 17 ± 1.5, and 16 ± 1.0, respectively. The IC50 value for NiHL1 after 24, 48, and 72 hours was 104 ± 3.5, 61 ± 4.0, and 48 ± 2.0, respectively.
Similarly, the IC50 values of utilized chemotherapeutic agents were evaluated by the MTT assay. H2O was used as the solvent for the drugs and their combination. Table 1 provides the prepared concentrations of applied drugs, such as cyclophosphamide (CP), methotrexate (MTX), mercaptopurine, cytarabine, methotrexate and cyclophosphamide (MTX + CP), cyclophosphamide and cytarabine (CP + Arac), cytarabine and mercaptopurine (Arac + 6 MP), and thiosemicarbazone Ni/Cu complexes. The prepared concentrations were tested in three time groups of 24, 48, and 72 hours, with drug-free cells serving as controls.
Table 1.
Drug Complex Formula
|
Complex
|
Drug 1
|
Drug 2
|
| Complex 1 |
1 µM MTX |
20 µM CP |
| Complex 2 |
20 µM CP |
1 µM cytarabine |
| Complex 3 |
1 µM cytarabine |
5 µM 6 MP |
Note. MTX: Methotrexate; CP: Cyclophosphamide; MP: Mercaptopurine.
Ribonucleic Acid Extraction, Complementary DNA Synthesis, and Real-Time Polymerase Chain Reaction
After passing the cells through the specified periods (24, 48, and 72) to assess the expression of the required genes, RNA was extracted using a total RNA extraction kit (PARS Company, Mashhad, Iran; CAT No. A101231). Following drug treatment, the extracted RNA was quantitatively purified using a spectrophotometer. The 260–280 nm ratio was in the 1.8–2.2 range, indicating no protein contamination, and the quantity of pure RNA was determined to be 0.5–1 g. Then, cDNA was synthesized from the acquired RNA using the Easy cDNA Reverse Transcription kit (50 tests Pars Tous CAT. No. A101161) following the kit’s instructions.
In RT-PCR, the necessary chemicals for synthesis were combined in a final volume of 10 μL Buffer 2x, 2 μL enzyme, 5 μL RNA, 3 μL ddH2O, and 3 μL total RNA and cycled for 15 minutes at 37 °C and then 8–16 seconds at 85 °C. Using a Rotor-Gene Q RT-PCR cycler (Qiagen, Hilden, Germany) and the Ampliqon SYBR green Master Mix kit (High ROX, Kalazist, No. A325402), the expression levels of ANRIL, CCAT1, NEAT1, and GAS5 lncRNAs, as well as GAPDH as a reference gene, were evaluated by PCR.
The primer sequences (5ʹ-3ʹ) used for ANRIL, CCAT1, NEAT1, and GAS5 lncRNAs were as follows:
Forward primer ANRIL (GGGCCTCAGTGGC ACATACC) and reverse primer (TGCTCTAT CCGCCAATCAGG), forward primer CCAT1 (TTTATGCTTGAGCCTTGA) and reverse primer (CTTGCCTGAAATACTTGC), forward primer GAS5 (CACACAGGCATTAGACAGA), and reverse primer (GCTCCACACAGTGTAGTCA).
A forward primer for NEAT1 (TGGCTAG CTCAGGGCTTCAG) and reverse primer (TCTCCT TGCCAAGCTTCCTTC), as well as GAPDH forward primer (ACCACA GTC CAT GCC ATC) and reverse primer (TCC ACC ACC CTG TTG CTG TA) were used as well. The FASTA sequence of the cDNA library of the genes was extracted from the National Center for Biotechnology Information (NCBI) website. The primer design was conducted using Primer3 software. To validate specificity and primer binding to genes, the data were blasted on the NCBI website using the aforementioned program.
The reactions were adjusted to a volume of 20 µL, combining 10 µL of Cyber Green (1X), 1 µL of forward and reverse primers (0.4 M), 2 µL of cDNA, and 6 µL of deionized water. The initial denaturation schedule was set at 95 °C for 15 minutes, and DNA fragments were amplified in 40 cycles, including denaturation at 95 °C for 5 seconds, annealing at 55 °C for 30 seconds (65 °C for lncRNA PCAT1), and extension at 72 °C for 30 seconds.
Gene Expression Confirmation and Statistical Analysis
To validate gene presence, the PCR product was electrophoresed on a 2% agarose gel, and the length of the fractions was equal to 300 bp. Subsequently, the PCR products were sequenced directly and authorized by Fanavaran Gene Company. Δct ct was determined for all cases and control samples by calculating the CT value of the target gene relative to the reference gene. A 2-ΔΔct (the fold change) was obtained for each sample. The Livak technique and the Rest software (2002) were utilized to assess the expression of lncRNA ANRIL, CCAT1, NEAT1, and GAS5 by RT-PCR. The significance threshold was set at P < 0.05.
Results
Effect of Methotrexate on Antisense Non-coding RNA in the INK4 Locus Expression
MTX treatment had a significant impact on the expression of ANRIL, a lncRNA. At a concentration of 1 µM, MTX induced a 2.21-fold increase in ANRIL expression after 24 hours of treatment. However, at a higher concentration of 10 µM, ANRIL expression was reduced by 0.584-fold at the same time point. Prolonged treatment for 48 hours and 72 hours also exhibited concentration-dependent effects on ANRIL expression, with decreasing fold changes observed at both concentrations (Figure 1).
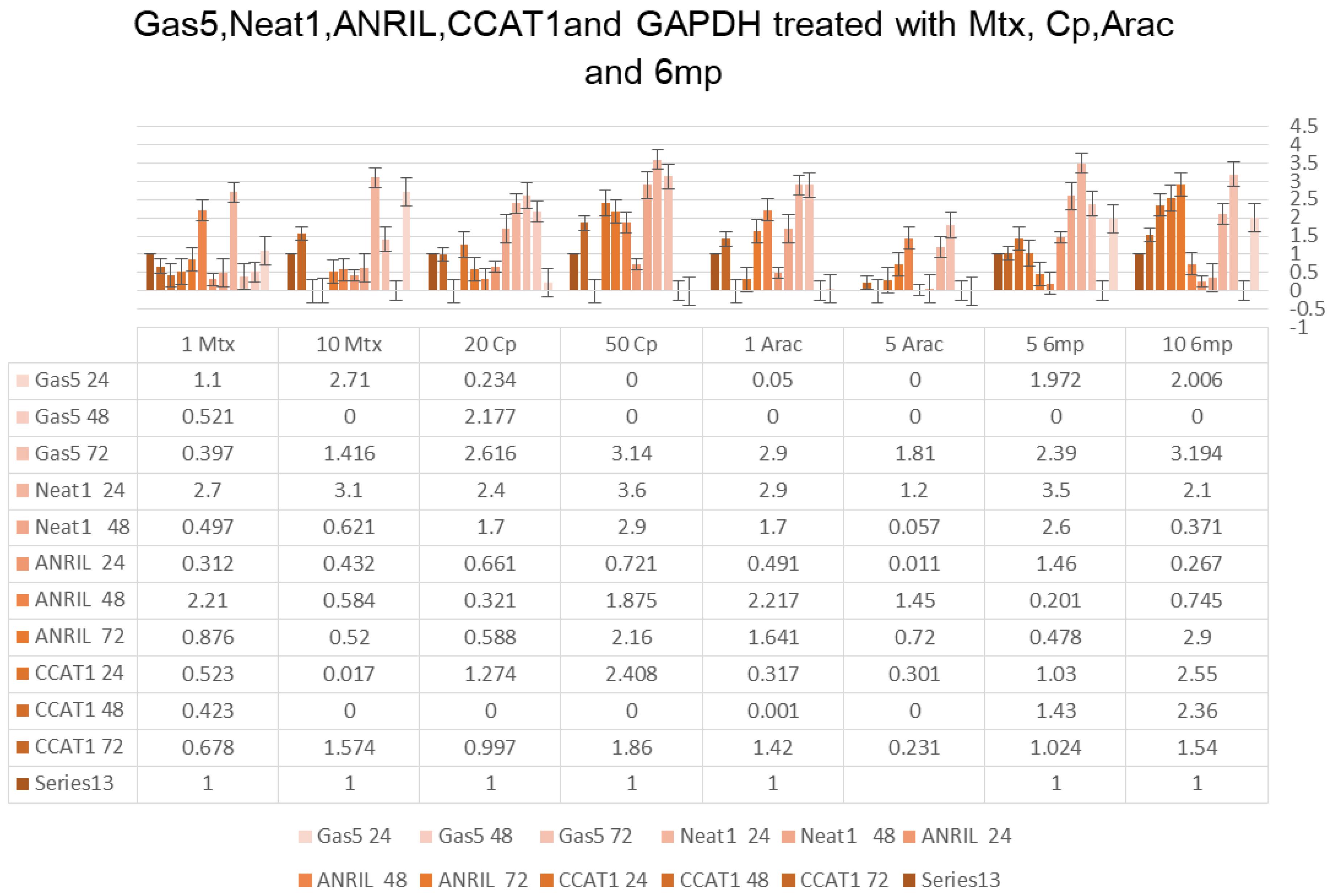
Figure 1.
GAS5, NEAT1, ANRIL, CCAT1, and GAPDH Treated With MTX, CP, Arac, and 6 MP. Note. MTX: Methotrexate; CP: Cyclophosphamide; MP: Mercaptopurine; GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
.
GAS5, NEAT1, ANRIL, CCAT1, and GAPDH Treated With MTX, CP, Arac, and 6 MP. Note. MTX: Methotrexate; CP: Cyclophosphamide; MP: Mercaptopurine; GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
Influence of Cyclophosphamide on Antisense Non-Coding RNA in the INK4 Locus Expression
CP treatment demonstrated significant effects on ANRIL expression. At a concentration of 20 µM, ANRIL expression was reduced by 0.321-fold after 24 hours, while treatment with 50 µM concentration resulted in a 1.875-fold increase at the same time point. Prolonged treatment durations of 48 hours and 72 hours showed similar concentration-dependent effects on ANRIL expression, with varying fold changes (Figure 1).
Impact of Cytarabine and Thiopurine Drugs on Antisense Non-coding RNA in the INK4 Locus Expression
Cytarabine (Arac) treatment led to altered ANRIL expression. At a concentration of 1 µM, ANRIL expression increased by 2.217-fold after 24 hours, while treatment with the 5 µM concentration resulted in a 1.45-fold increase. However, ANRIL expression decreased with increasing concentrations of cytarabine and longer treatment durations. Thiopurine drugs (6 MP) also represented concentration-dependent effects on ANRIL expression, with varying fold changes observed at different concentrations and time points (Figure 1).
Influence of Thiosemicarbazone (Ni and Cu) on Antisense Non-coding RNA in the INK4 Locus Expression
Treatment with thiosemicarbazone (Ni) and thiosemicarbazone (Cu) influenced ANRIL expression. Thiosemicarbazone (Ni) treatment at the 100.5 µM concentration could significantly decrease ANRIL expression after 24 hours, while treatment with the 104 µM concentration revealed no change. However, after 72 hours, higher concentrations of thiosemicarbazone (Ni) induced higher ANRIL expression. Thiosemicarbazone (Cu) treatment also exhibited a similar trend, with no effect on ANRIL expression after 24 hours but a significant increase after 72 hours (Figure 2).
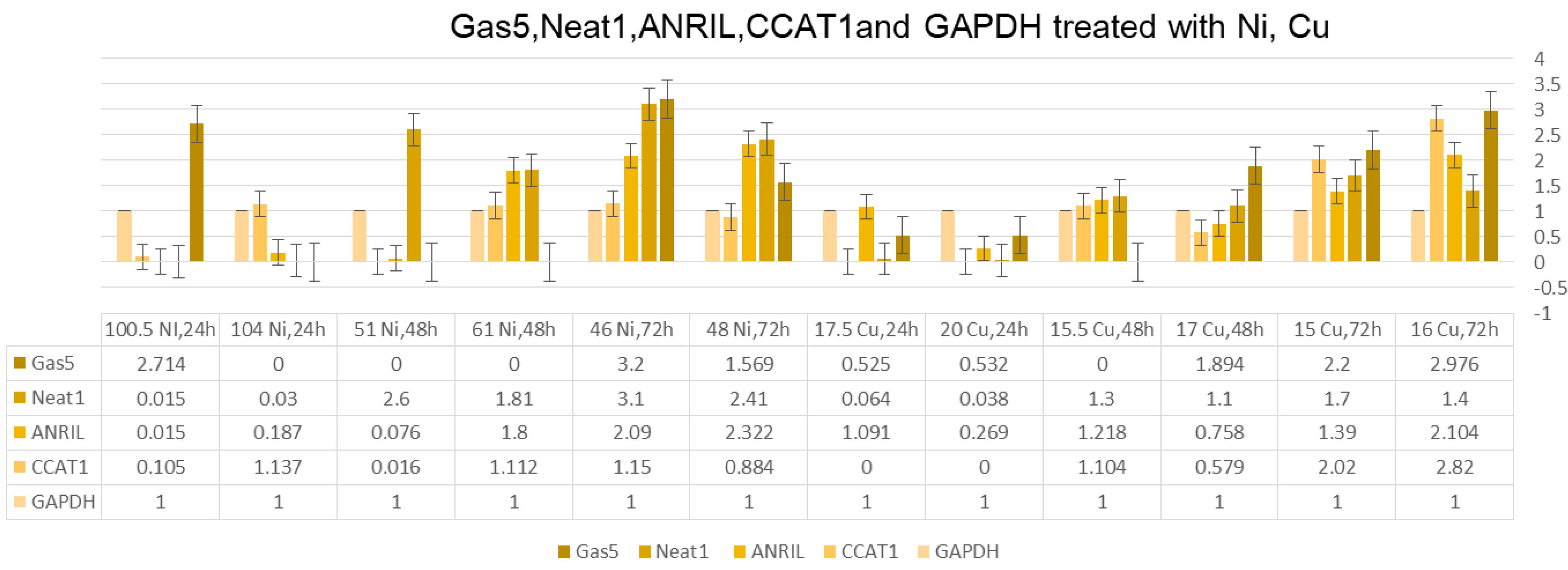
Figure 2.
GAS5, NEAT1, ANRIL, CCAT1, and GAPDH Treated With Ni and Cu. Note. GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
.
GAS5, NEAT1, ANRIL, CCAT1, and GAPDH Treated With Ni and Cu. Note. GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
Expression of Antisense Non-coding RNA in the INK4 Locus in Drug Combinations
The combination treatments, including Complex 1 (1 µM MTX + 20 µM CP), Complex 2 (20 µM CP + 1 µm cytarabine), and Complex 3 (1 µM cytarabine + 5 µM 6 MP), also influenced ANRIL expression. Complex 1 treatment showed a 0.321-fold decrease in ANRIL expression after 24 hours. Similarly, Complexes 2 and 3 had distinct effects on ANRIL expression, with both combinations demonstrating variable fold changes after 24 hours, 48 hours, and 72 hours of treatment (Figure 3).
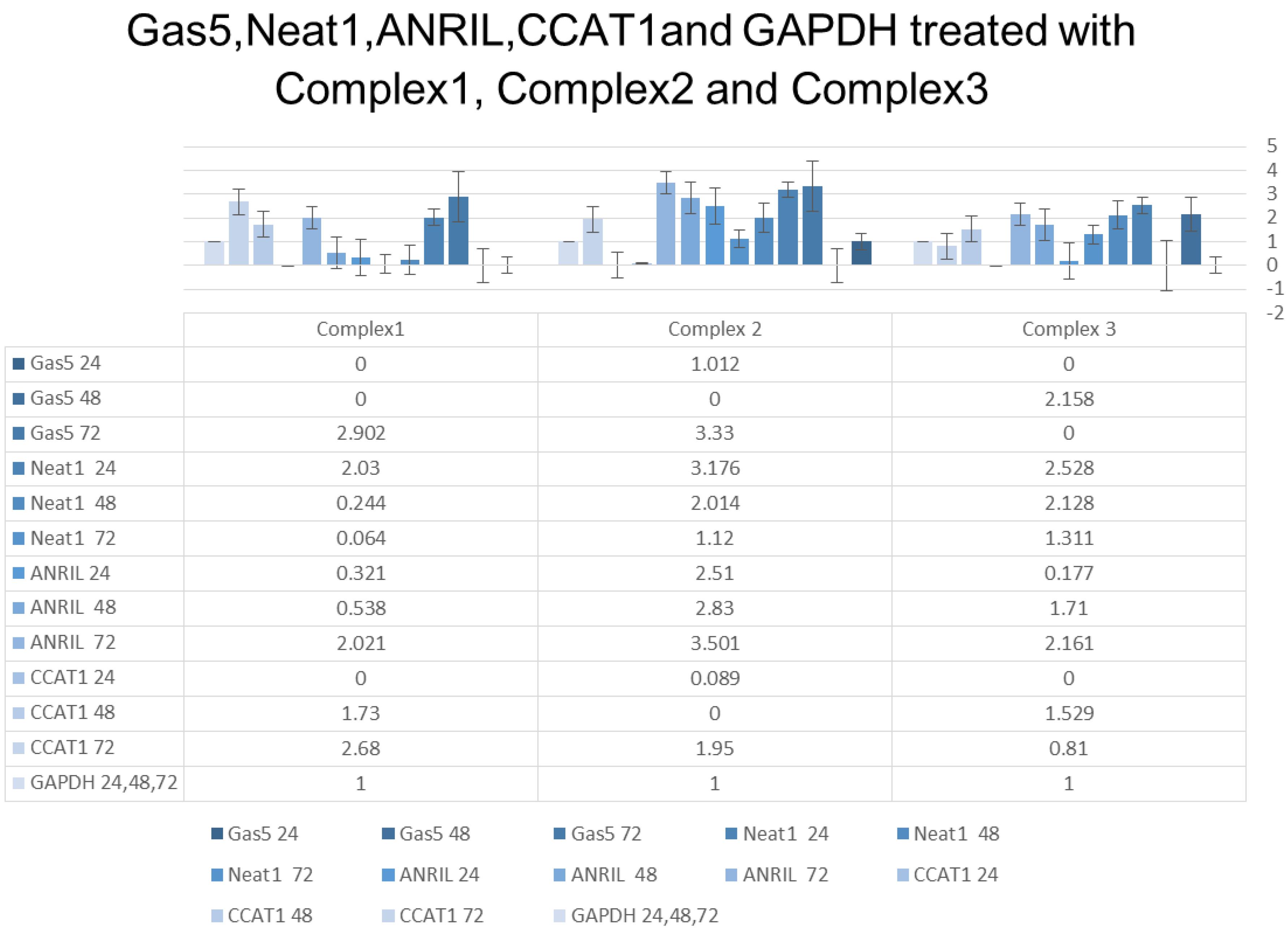
Figure 3.
GAS5, NEAT1, ANRIL, CCAT1, and GAPDH Treated With Complex 1, Complex 2, and Complex 3. Note. GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
.
GAS5, NEAT1, ANRIL, CCAT1, and GAPDH Treated With Complex 1, Complex 2, and Complex 3. Note. GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
Time-Dependent Effects on Antisense Non-coding RNA in the INK4 Locus Expression
The duration of drug treatment played a crucial role in regulating ANRIL expression. ANRIL expression represented time-dependent changes, with varying fold changes found at different treatment durations for each drug and drug combination tested.
Effect of Methotrexate on Colon Cancer-Associated Transcript 1 Expression
Similar to ANRIL, the expression of CCAT1, another lncRNA, was also influenced by MTX treatment. At a concentration of 1 µM, CCAT1 expression was reduced by 0.423-fold after 24 hours, while no expression was detected at a higher concentration of 10 µM. Prolonged treatment for 48 hours and 72 hours also revealed concentration-dependent effects on CCAT1 expression (Figure 1).
Influence of Cyclophosphamide Carboplatin on Colon Cancer-Associated Transcript 1 Expression
Cyclophosphamide treatment had varied effects on CCAT1 expression. While no expression of CCAT1 was observed after 24 hours of treatment at both 20 µM and 50 µM concentrations, treatment for 48 hours and 72 hours resulted in differential fold changes in CCAT1 expression (Figure 1).
Impact of Cytarabine and Thiopurine Drugs on Colon Cancer-Associated Transcript 1 Expression
Cytarabine treatment exhibited concentration-dependent effects on CCAT1 expression, with varying fold changes observed at different concentrations and time points. Thiopurine drugs (6 MP) also affected CCAT1 expression, showing both upregulation and downregulation at different concentrations and treatment durations (Figure 1).
Expression of Colon Cancer-Associated Transcript 1 in Drug Combinations
The combination treatments, including Complex 1 (1 µM MTX + 20 µM CP), Complex 2 (20 µM CP + 1 µM cytarabine), and Complex 3 (1 µM cytarabine + 5 µM 6 MP), represented distinct effects on CCAT1 expression. Complex 1 treatment resulted in a 0.321-fold decrease in CCAT1 expression after 24 hours. Complexes 2 and 3 demonstrated diverse fold changes in CCAT1 expression at different time points (Figure 3).
These results confirmed the significant influence of various anti-cancer drugs, both individually and in combination, on the expression of ANRIL and CCAT1, highlighting the importance of these lncRNAs in the context of cancer treatment. Further investigations are warranted to elucidate the precise molecular mechanisms underlying these observed effects and to explore the potential clinical implications of targeting ANRIL and CCAT1 in cancer therapy.
Effect of Methotrexate on Nuclear Enriched Abundant Transcript 1 and Growth Arrest-Specific 5 Expression
MTX treatment had considerable effects on the expression of NEAT1 and GAS5, two lncRNAs. Treatment with 1 µM MTX for 24 hours caused a 2.7-fold increase in NEAT1 expression and a 1.1-fold increase in GAS5 expression. However, a higher concentration of 10 µM MTX showed a more pronounced effect, leading to a 3.1-fold increase in NEAT1 expression and a 2.71-fold increase in GAS5 expression. Prolonged treatment durations of 48 hours and 72 hours revealed concentration-dependent effects on NEAT1 and GAS5 expression, with varying fold changes (Figure 1).
Influence of Cyclophosphamide on Nuclear Enriched Abundant Transcript 1 and Growth Arrest-Specific 5 Expression
Cyclophosphamide treatment also exhibited significant impacts on NEAT1 and GAS5 expression. Treatment with 20 µM CP for 24 hours resulted in a 2.4-fold increase in NEAT1 expression but no change in GAS5 expression. Conversely, treatment with 50 µM CP for the same duration confirmed a more substantial effect, leading to a 3.6-fold increase in NEAT1 expression but no GAS5 expression. Prolonged treatment durations of 48 hours and 72 hours demonstrated variable fold changes in NEAT1 and GAS5 expression (Figure 1).
Impact of Cytarabine and Thiopurine Drugs on Nuclear Enriched Abundant Transcript 1 and Growth Arrest-Specific 5 Expression
Cytarabine (Arac) treatment represented concentration-dependent effects on NEAT1 and GAS5 expression. Treatment with 1 µM Arac for 24 hours led to a 2.9-fold increase in NEAT1 expression, while no GAS5 expression was detected. In contrast, treatment with 5 µM Arac for the same duration caused no NEAT1 or GAS5 expression. Prolonged treatment durations and higher concentrations of Arac showed no NEAT1 or GAS5 expression.
Thiopurine drugs (6 MP) also influenced NEAT1 and GAS5 expression. Treatment with 5 µM 6 MP for 24 hours resulted in 3.5-fold and 1.972-fold increases in NEAT1 and GAS5 expression, respectively. Similarly, treatment with 10 µM 6 MP for the same duration led to a 2.1-fold increase in NEAT1 expression and a 2.006-fold increase in GAS5 expression. Prolonged treatment durations of 48 hours and 72 hours indicated varying fold changes in NEAT1 and GAS5 expression (Figure 1).
Influence of Thiosemicarbazone (Ni and Cu) on Nuclear Enriched Abundant Transcript 1 and Growth Arrest-Specific 5 Expression
Treatment with thiosemicarbazone (Ni) and thiosemicarbazone (Cu) had distinct effects on NEAT1 and GAS5 expression. Thiosemicarbazone (Ni) treatment at the 100.5 µM concentration resulted in a 2.714-fold increase in NEAT1 expression after 24 hours, while treatment with the 104 µM concentration demonstrated no NEAT1 expression. For GAS5, no expression was detected for both concentrations after 24 hours. Prolonged treatment for 48 hours and 72 hours exhibited variable fold changes in NEAT1 and GAS5 expression (Figure 2).
Expression of Nuclear Enriched Abundant Transcript 1 and Growth Arrest-Specific 5 in Drug Combinations
The combination treatments, including Complex 1 (1 µM MTX + 20 µM CP), Complex 2 (20 µM CP + 1 µM cytarabine), and Complex 3 (1 µM cytarabine + 5 µM 6 MP), revealed diverse effects on NEAT1 and GAS5 expression. Complex 1 treatment led to a 2.03-fold increase in NEAT1 expression, but no GAS5 expression was detected after 24 hours. Complex 2 represented a 3.176-fold increase in NEAT1 expression and a 1.012-fold increase in GAS5 expression for the same duration. Complex 3 demonstrated a 2.528-fold increase in NEAT1 expression, while no GAS5 expression was detected after 24 hours. Prolonged treatment durations of 48 hours and 72 hours showed various fold changes in NEAT1 and GAS5 expression for these drug combinations (Figure 3).
Discussion
The findings presented in this study shed light on the impact of thiosemicarbazone complexes and various chemotherapy drugs on the expression profiles of lncRNAs ANRIL, CCAT1, NEAT1, and GAS5 in the Jurkat E6.1 ALL cell line. The results revealed a complex interplay between these therapeutic agents and lncRNA expression, providing insights into potential mechanisms underlying their effects and highlighting the importance of these lncRNAs in cancer biology. MTX, a commonly used chemotherapy drug, had significant effects on the expression of ANRIL and CCAT1. Interestingly, MTX treatment exhibited contrasting effects on the two lncRNAs, with ANRIL expression showing a concentration-dependent increase while CCAT1 expression was reduced. These findings suggest distinct regulatory mechanisms for ANRIL and CCAT1 in response to MTX treatment. CP treatment also demonstrated notable effects on ANRIL and CCAT1 expression. While CP did not induce CCAT1 expression, it led to increased ANRIL expression, indicating differential regulation of these lncRNAs by CP. These findings emphasize the complexity of lncRNA regulation in response to specific chemotherapy agents. The impact of cytarabine and thiopurine drugs on ANRIL and CCAT1 expression also underwent investigation. Cytarabine treatment displayed concentration-dependent effects on ANRIL expression, with varying fold changes observed at different concentrations and time points. Similarly, thiopurine drugs had concentration-dependent effects on both ANRIL and CCAT1 expression, demonstrating the potential interplay between these drugs and lncRNA regulation. Furthermore, the combination treatments involving multiple drugs (Complex 1: MTX + CP, Complex 2: CP + Cytarabine, Complex 3: Cytarabine + 6 MP) revealed distinct effects on ANRIL and CCAT1 expression profiles. These combinations resulted in diverse fold changes, suggesting potential synergistic or antagonistic interactions between the drugs in modulating lncRNA expression. It is worth noting that the duration of drug treatment played a crucial role in the regulation of ANRIL and CCAT1 expression. Time-dependent changes in lncRNA expression were observed, emphasizing the importance of considering treatment duration when evaluating the effects of chemotherapy agents on lncRNAs.
ANRIL, located in the INK4 locus on chromosome 9p21,9 has garnered significant attention due to its association with multiple cancer types.10 ANRIL exerts its oncogenic effects by modulating cell cycle progression, apoptosis, and DNA repair pathways. High ANRIL expression has been observed in several cancers, including melanoma,11 glioblastoma,12 prostate,13 and pancreatic cancer.14 ANRIL’s involvement in cancer is often attributed to its ability to silence tumor suppressor genes, such as p15INK4B, p16INK4A, and ARF, through epigenetic modifications.15 Moreover, ANRIL is implicated in promoting angiogenesis, epithelial-mesenchymal transition, and metastasis, making it a promising therapeutic target.14 CCAT1, originally identified in colorectal cancer, has since been implicated in several malignancies.16 CCAT1 exhibits elevated expression levels in various cancers, including gastric,17 breast,18 lung,19 and hepatocellular carcinomas.20 This lncRNA contributes to cancer development by promoting the proliferation, invasion, and migration of cancer cells. CCAT1 achieves these effects by interacting with chromatin remodeling factors and transcription factors, ultimately modulating gene expression.21 Notably, CCAT1 has also been associated with resistance to chemotherapy and radiotherapy, highlighting its potential role in therapeutic resistance.22,23
NEAT1, primarily localized in nuclear paraspeckles, is involved in multiple cellular processes, including cancer progression.24 Its dysregulation has been observed in various malignancies, such as breast,25 lung,26 and ovarian cancer.27 NEAT1 has been implicated in promoting tumor growth, metastasis, and drug resistance.28 Mechanistically, NEAT1 interacts with oncogenic factors, including epigenetic regulators, miRNAs, and transcription factors, thereby affecting gene expression patterns.29 NEAT1’s association with poor prognosis and therapeutic resistance underscores its potential as a prognostic marker and therapeutic target.30
GAS5, unlike the previously mentioned lncRNAs, functions as a tumor suppressor in cancer.31 GAS5 expression is frequently downregulated in multiple cancers, including breast, lung, and prostate cancer.32 GAS5 exerts its tumor-suppressive effects by regulating cell cycle progression, apoptosis, and cellular senescence.33 It achieves this by sequestering and inhibiting the activity of various oncogenic proteins, including glucocorticoid receptors and miRNAs. The reintroduction or upregulation of GAS5 has shown promising results in suppressing tumor growth and sensitizing cancer cells to chemotherapy.
The observed effects on lncRNA expression profiles provide valuable insights into the potential roles of ANRIL and CCAT1 in cancer and the therapeutic response to these drugs. ANRIL has been implicated in various cancer types and has been associated with cell proliferation, apoptosis, and angiogenesis. CCAT1 has also been linked to cancer progression, metastasis, and drug resistance. Therefore, the altered expression of these lncRNAs in response to chemotherapy agents suggests their potential as biomarkers or therapeutic targets in leukemia treatment.
However, it is noteworthy that the mechanisms underlying the observed effects on lncRNA expression remain to be elucidated. Further studies are warranted to investigate the precise molecular pathways involved in the regulation of ANRIL and CCAT1 by these drugs. Additionally, functional studies exploring the downstream effects of altered lncRNA expression are essential to fully understand their roles in cancer biology and their potential as therapeutic targets.
Suggestion and Implication
Ultimately, it is hoped that the findings contribute to the ongoing efforts to improve treatment outcomes and advance the field of precision medicine in ALL. Further investigations are warranted to unravel the functional significance of these lncRNAs and their intricate interactions with the cellular pathways involved in ALL pathogenesis. Integrating multi-omics approaches, such as RNA sequencing and functional assays, could provide a more comprehensive understanding of the molecular landscape of ALL and the role of lncRNAs within it. Additionally, in vivo studies utilizing relevant animal models and patient-derived samples would strengthen the translational relevance of the findings.
Conclusion
This study provides valuable insights into the effects of thiosemicarbazone complexes and chemotherapy drugs on the expression profiles of ANRIL, CCAT1, NEAT1, and GAS5 in ALL cells. The findings underscore the complex interplay between these therapeutic agents and lncRNA expression, highlighting the potential roles of ANRIL and CCAT1 as key players in leukemia biology and treatment response. Further investigations are warranted to unravel the underlying molecular mechanisms and explore the clinical implications of targeting these lncRNAs in leukemia therapy. In conclusion, this study provides valuable insights into the effects of thiosemicarbazone complexes and chemotherapy drugs on the expression profiles of ANRIL, CCAT1, NEAT1, and GAS5 in ALL cells. The findings underscore the complex interplay between these therapeutic agents and lncRNA expression, highlighting the potential roles of ANRIL and CCAT1 as key players in leukemia biology and treatment response. Further investigations are warranted to unravel the underlying molecular mechanisms and explore the clinical implications of targeting these lncRNAs in leukemia therapy (Figure 4).
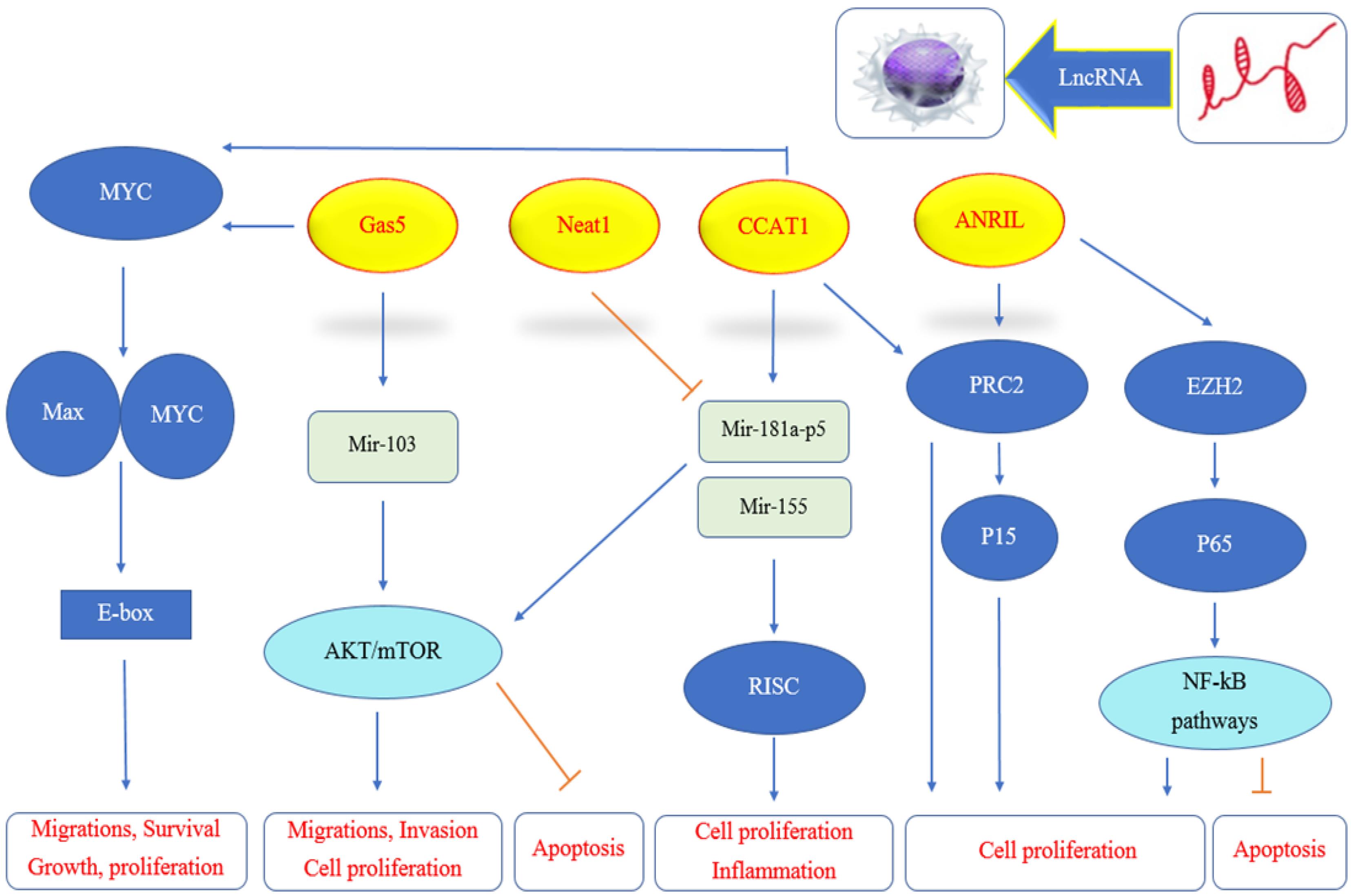
Figure 4.
Summery of LncRNA effect on immunity. Note. GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; LncRNA: Long non-coding RNA
.
Summery of LncRNA effect on immunity. Note. GAS5: Growth arrest-specific 5; NEAT1: Nuclear enriched abundant transcript 1; ANRIL: Antisense non-coding RNA in the INK4 locus; CCAT1: Colon cancer-associated transcript 1; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; LncRNA: Long non-coding RNA
Authors’ Contribution
Conceptualization: Sina Mirzaahmadi.
Data curation: Sina Mirzaahmadi.
Formal analysis: Sina Mirzaahmadi.
Investigation: Neda Zahmatkesh.
Methodology: Neda Zahmatkesh.
Project administration: Golnaz Asaadi Tehrani.
Resources: Neda Zahmatkesh.
Software: Saedeh Hatampour.
Supervision: Golnaz Asaadi Tehrani.
Validation: Saedeh Hatampour.
Visualization: Mahsa Qaed.
Writing–original draft: Mahsa Qaed and Saedeh Hatampour.
Writing–review & editing: Golnaz Asaadi Tehrani.
Competing Interests
The authors declare that they have no competing interests.
Ethical Approval
This study received approval from the Ethics Committee of Islamic Azad University, Zanjan Branch, Zanjan, Iran (ethical code IR.IAU.Z.REC.1401.038).
References
- Lejman M, Chałupnik A, Chilimoniuk Z, Dobosz M. Genetic biomarkers and their clinical implications in B-cell acute lymphoblastic leukemia in children. Int J Mol Sci 2022; 23(5):2755. doi: 10.3390/ijms23052755 [Crossref] [ Google Scholar]
- Esparza SD, Sakamoto KM. Topics in pediatric leukemia--acute lymphoblastic leukemia. MedGenMed 2005; 7(1):23. [ Google Scholar]
- Zhou Z, Lin Z, Pang X, Tariq MA, Ao X, Li P. Epigenetic regulation of long non-coding RNAs in gastric cancer. Oncotarget 2018; 9(27):19443-58. doi: 10.18632/oncotarget.23821 [Crossref] [ Google Scholar]
- Shahmir S, Zahmatkesh N, Mirzaahmadi S, Asaadi Tehrani G. LncRNA CASC2 inhibits progression of glioblastoma by regulating the expression of AKT in T98G cell line, treated by TMZ and thiosemicarbazone complex. Asian Pac J Cancer Prev 2023; 24(5):1553-60. doi: 10.31557/apjcp.2023.24.5.1553 [Crossref] [ Google Scholar]
- Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci 2015; 16(2):3251-66. doi: 10.3390/ijms16023251 [Crossref] [ Google Scholar]
- Di Martino MT, Riillo C, Scionti F, Grillone K, Polerà N, Caracciolo D. miRNAs and lncRNAs as novel therapeutic targets to improve cancer immunotherapy. Cancers (Basel) 2021; 13(7):1587. doi: 10.3390/cancers13071587 [Crossref] [ Google Scholar]
- Hassani A, Asaadi Tehrani G, Zahmatkesh N, Mirzaahmadi S. Evaluation the effect of chemotherapy drugs and thiosemicarbazone complexes on the expression of URHC and CASC15 LncRNAs in acute lymphoblastic leukemia cell line. Iran J Blood Cancer 2023; 15(1):1-9. doi: 10.58209/ijbc.15.1.1 [Crossref] [ Google Scholar]
- Guo ZL, Richardson DR, Kalinowski DS, Kovacevic Z, Tan-Un KC, Chan GC. The novel thiosemicarbazone, di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC), inhibits neuroblastoma growth in vitro and in vivo via multiple mechanisms. J Hematol Oncol 2016; 9(1):98. doi: 10.1186/s13045-016-0330-x [Crossref] [ Google Scholar]
- Razeghian-Jahromi I, Karimi Akhormeh A, Zibaeenezhad MJ. The role of ANRIL in atherosclerosis. Dis Markers 2022; 2022:8859677. doi: 10.1155/2022/8859677 [Crossref] [ Google Scholar]
- Li Z, Yu X, Shen J. ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol 2016; 37(5):5657-61. doi: 10.1007/s13277-016-4808-5 [Crossref] [ Google Scholar]
- Sarkar D, Oghabian A, Bodiyabadu PK, Joseph WR, Leung EY, Finlay GJ. Multiple isoforms of ANRIL in melanoma cells: structural complexity suggests variations in processing. Int J Mol Sci 2017; 18(7):1378. doi: 10.3390/ijms18071378 [Crossref] [ Google Scholar]
- Sun Y, Jing Y, Zhang Y. Serum lncRNA-ANRIL and SOX9 expression levels in glioma patients and their relationship with poor prognosis. World J Surg Oncol 2021; 19(1):287. doi: 10.1186/s12957-021-02392-2 [Crossref] [ Google Scholar]
- Taheri M, Pouresmaeili F, Omrani MD, Habibi M, Sarrafzadeh S, Noroozi R. Association of ANRIL gene polymorphisms with prostate cancer and benign prostatic hyperplasia in an Iranian population. Biomark Med 2017; 11(5):413-22. doi: 10.2217/bmm-2016-0378 [Crossref] [ Google Scholar]
- Chen S, Zhang JQ, Chen JZ, Chen HX, Qiu FN, Yan ML. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: an in vivo and in vitro study. Int J Biol Macromol 2017; 102:718-28. doi: 10.1016/j.ijbiomac.2017.03.123 [Crossref] [ Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011; 30(16):1956-62. doi: 10.1038/onc.2010.568 [Crossref] [ Google Scholar]
- Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer 2013; 13:196. doi: 10.1186/1471-2407-13-196 [Crossref] [ Google Scholar]
- Zhu H, Zhao H, Zhang L, Xu J, Zhu C, Zhao H. Dandelion root extract suppressed gastric cancer cells proliferation and migration through targeting lncRNA-CCAT1. Biomed Pharmacother 2017; 93:1010-7. doi: 10.1016/j.biopha.2017.07.007 [Crossref] [ Google Scholar]
- Selem NA, Youness RA, Gad MZ. What is beyond LncRNAs in breast cancer: a special focus on colon cancer-associated Transcript-1 (CCAT-1). Noncoding RNA Res 2021; 6(4):174-86. doi: 10.1016/j.ncrna.2021.11.001 [Crossref] [ Google Scholar]
- Chen J, Alduais Y, Zhang K, Zhu X, Chen B. CCAT1/FABP5 promotes tumour progression through mediating fatty acid metabolism and stabilizing PI3K/AKT/mTOR signalling in lung adenocarcinoma. J Cell Mol Med 2021; 25(19):9199-213. doi: 10.1111/jcmm.16815 [Crossref] [ Google Scholar]
- Guo X, Hua Y. CCAT1: an oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol 2017; 143(4):555-62. doi: 10.1007/s00432-016-2268-3 [Crossref] [ Google Scholar]
- Huang Y, Guo Q, Ding XP, Wang X. Mechanism of long noncoding RNAs as transcriptional regulators in cancer. RNA Biol 2020; 17(11):1680-92. doi: 10.1080/15476286.2019.1710405 [Crossref] [ Google Scholar]
- Li X, Han X, Wei P, Yang J, Sun J. Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol Ther 2020; 21(5):452-62. doi: 10.1080/15384047.2020.1727700 [Crossref] [ Google Scholar]
- Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int 2018; 42(2):227-36. doi: 10.1002/cbin.10890 [Crossref] [ Google Scholar]
- Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol 2019; 13(1):46-60. doi: 10.1002/1878-0261.12404 [Crossref] [ Google Scholar]
- Knutsen E, Harris AL, Perander M. Expression and functions of long non-coding RNA NEAT1 and isoforms in breast cancer. Br J Cancer 2022; 126(4):551-61. doi: 10.1038/s41416-021-01588-3 [Crossref] [ Google Scholar]
- Jiang P, Wu X, Wang X, Huang W, Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016; 7(28):43337-51. doi: 10.18632/oncotarget.9712 [Crossref] [ Google Scholar]
- Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci 2016; 20(16):3373-7. [ Google Scholar]
- Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW, Wang X. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis 2019; 10(4):270. doi: 10.1038/s41419-019-1513-5 [Crossref] [ Google Scholar]
- Kumar S, Gonzalez EA, Rameshwar P, Etchegaray JP. Non-coding RNAs as mediators of epigenetic changes in malignancies. Cancers (Basel) 2020; 12(12):3657. doi: 10.3390/cancers12123657 [Crossref] [ Google Scholar]
- Thankachan S, Bhardwaj BK, Venkatesh T, Suresh PS. Long non-coding RNA NEAT1 as an emerging biomarker in breast and gynecologic cancers: a systematic overview. Reprod Sci 2021; 28(9):2436-47. doi: 10.1007/s43032-021-00481-x [Crossref] [ Google Scholar]
- Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol 2016; 37(2):1437-44. doi: 10.1007/s13277-015-4521-9 [Crossref] [ Google Scholar]
- Yang X, Xie Z, Lei X, Gan R. Long non-coding RNA GAS5 in human cancer. Oncol Lett 2020; 20(3):2587-94. doi: 10.3892/ol.2020.11809 [Crossref] [ Google Scholar]
- Heydarnezhad Asl M, Pasban Khelejani F, Bahojb Mahdavi SZ, Emrahi L, Jebelli A, Mokhtarzadeh A. The various regulatory functions of long noncoding RNAs in apoptosis, cell cycle, and cellular senescence. J Cell Biochem 2022; 123(6):995-1024. doi: 10.1002/jcb.30221 [Crossref] [ Google Scholar]